Today, I did computing of Chapter 10.
Codes:
library(AppliedPredictiveModeling)
data(concrete)
str(concrete)
str(mixtures)
library(Hmisc)
library(caret)
# g: grid; p: points; smooth: smoother between: add space between panels.
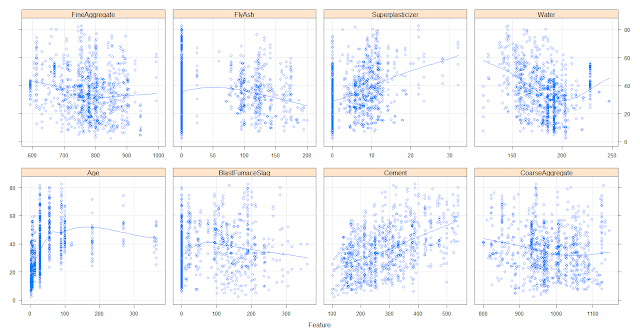
featurePlot(x=concrete[,-9], y=concrete$CompressiveStrength,
between=list(x=1, y=1),
type=c("g", "p", "smooth"))
# ? averaging the replicated mixtures and splitting the data into training and test sets
library(plyr)
averaged=ddply(mixtures, .(Cement, BlastFurnaceSlag, FlyAsh, Water, Superplasticizer, CoarseAggregate,
FineAggregate, Age), function(x) c(CompressiveStrength=mean(x$CompressiveStrength)))
str(averaged)
set.seed(975)
fortraining=createDataPartition(averaged$CompressiveStrength, p=3/4)[[1]]
trainingset=averaged[fortraining,]
testset=averaged[-fortraining,]
dim(trainingset)
dim(testset)
# The dot in the formula below is shorthand for all predictors and
# (.)^2 expands into a model with all the linear terms and all two-factor interactions.
modformula=paste("CompressiveStrength ~ (.)^2 + I(Cement^2) + ",
"I(BlastFurnaceSlag^2)+I(FlyAsh^2)+I(Water^2)+",
"I(Superplasticizer^2)+I(CoarseAggregate^2)+",
"I(FineAggregate^2)+I(Age^2)")
modformula=as.formula(modformula)
modformula
# the predictors' number of modformula is different from the book, but after the test of a different one, modformula2,
modformula2=paste("CompressiveStrength ~ (.)^2")
modformula2=as.formula(modformula2)
modformula2
# modformula and modformula2 have different results of linearreg although their predictors' numbers are the same.
# so I guess that modformula is correct.
controlobject=trainControl(method="repeatedcv", repeats=5, number=10)
set.seed(669)
linearreg=train(modformula, data=trainingset, method="lm", trControl=controlobject)
linearreg
# the other two linear models were created
set.seed(669)
plsmodel=train(modformula, data=trainingset, method="pls", preProcess = c("center", "scale"),
tuneLength = 15, trControl = controlobject)
plsmodel
# centered (44) and scaled (44): 44 is right.
library(elasticnet)
enetgrid2=expand.grid(.lambda=c(0, 0.001, 0.01, 0.1),
.fraction=seq(0.05, 1, length=20))
set.seed(669)
enetmodel2=train(modformula, data=trainingset, method="enet", preProcess = c("center", "scale"),
tuneGrid=enetgrid2, trControl=controlobject)
enetmodel2
# MARS
library(earth)
set.seed(669)
earthmodel=train(CompressiveStrength~., data=trainingset, method="earth",
tuneGrid=expand.grid(.degree=1, .nprune=2:25), trControl=controlobject)
earthmodel
#SVMs
library(kernlab)
set.seed(669)
svmmodel=train(CompressiveStrength~., data=trainingset, method="svmRadial",
tuneLength=15, preProcess=c("center", "scale"), trControl=controlobject)
svmmodel
# Neural Networks
library(nnet)
nnetgrid=expand.grid(.decay=c(0.001, 0.01, 0.1), .size=seq(1, 27, by=2), .bag=FALSE)
set.seed(669)
nnetmodel=train(CompressiveStrength~., data=trainingset, method="avNNet",
tuneGrid=nnetgrid, preProcess=c("center", "scale"),
linout=TRUE, trace=FALSE, maxit=1000, trControl=controlobject)
nnetmodel
# regression and model trees
library(rpart)
set.seed(669)
rpartmodel=train(CompressiveStrength~., data=trainingset, method="rpart",
tuneLength=30, trControl=controlobject)
rpartmodel
set.seed(669)
library(party)
ctreemodel=train(CompressiveStrength~., data=trainingset, method="ctree",
tuneLength=10, trControl=controlobject)
ctreemodel
set.seed(669)
library(RWeka)
mtmodel=train(CompressiveStrength~., data=trainingset, method="M5",
trControl=controlobject)
mtmodel
# remaining model objects
library(ipred)
library(plyr)
library(e1071)
set.seed(669)
treebagmodel=train(CompressiveStrength~., data=trainingset, method="treebag",
trControl=controlobject)
treebagmodel
library(randomForest)
set.seed(669)
rfmodel=train(CompressiveStrength~., data=trainingset, method="rf",
tuneLength=7, ntrees=1000, importance=TRUE,
trControl=controlobject)
rfmodel
gbmgrid=expand.grid(.interaction.depth = seq(1, 7, by = 2),
.n.trees = seq(100, 1000, by = 50),
.shrinkage = c(0.01, 0.1), .n.minobsinnode=10)
library(gbm)
set.seed(669)
gbmmodel=train(CompressiveStrength~., data=trainingset, method="gbm",
tuneGrid=gbmgrid, verbose=FALSE, trControl=controlobject)
gbmmodel
cubistgrid=expand.grid(.committees = c(1, 5, 10, 50, 75, 100),
.neighbors = c(0, 1, 3, 5, 7, 9))
library(Cubist)
set.seed(669)
cbmodel=train(CompressiveStrength~., data=trainingset, method="cubist",
tuneGrid=cubistgrid, trControl=controlobject)
cbmodel
allresamples=resamples(list("Linear Reg"=lmmodel), "PLS"=plsmodel,
"Elastic Net"=enetmodel2, "MARS"=earthmodel,
"SVM"=svmmodel,
# "Neural Networks"=nnetmodel,
"CART"=rpartmodel, "Cond Inf Tree" = ctreemodel,
"Bagged Tree" = treebagmodel, "Boosted Tree" = gbmmodel,
"Random Forest" = rfmodel, "Cubist" = cbmodel))
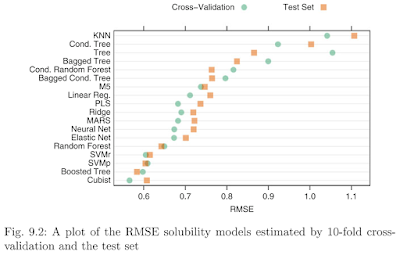
(There should be a figure of comparison among these models. However, the nnetmodel takes so much time to calculate that it has not shown the result till now. I will keep it calculating all the night and it should show the result tomorrow.)
Tomorrow, I will continue to do computing of Chapter 10.